Devin uses a PP polypropylene filter with pores of 15 nm that is coated with Zwitterions.
The zwitterion polypropylene is a superior and faster patented technology for eliminating lymphocytes that carry positive and negative charges.
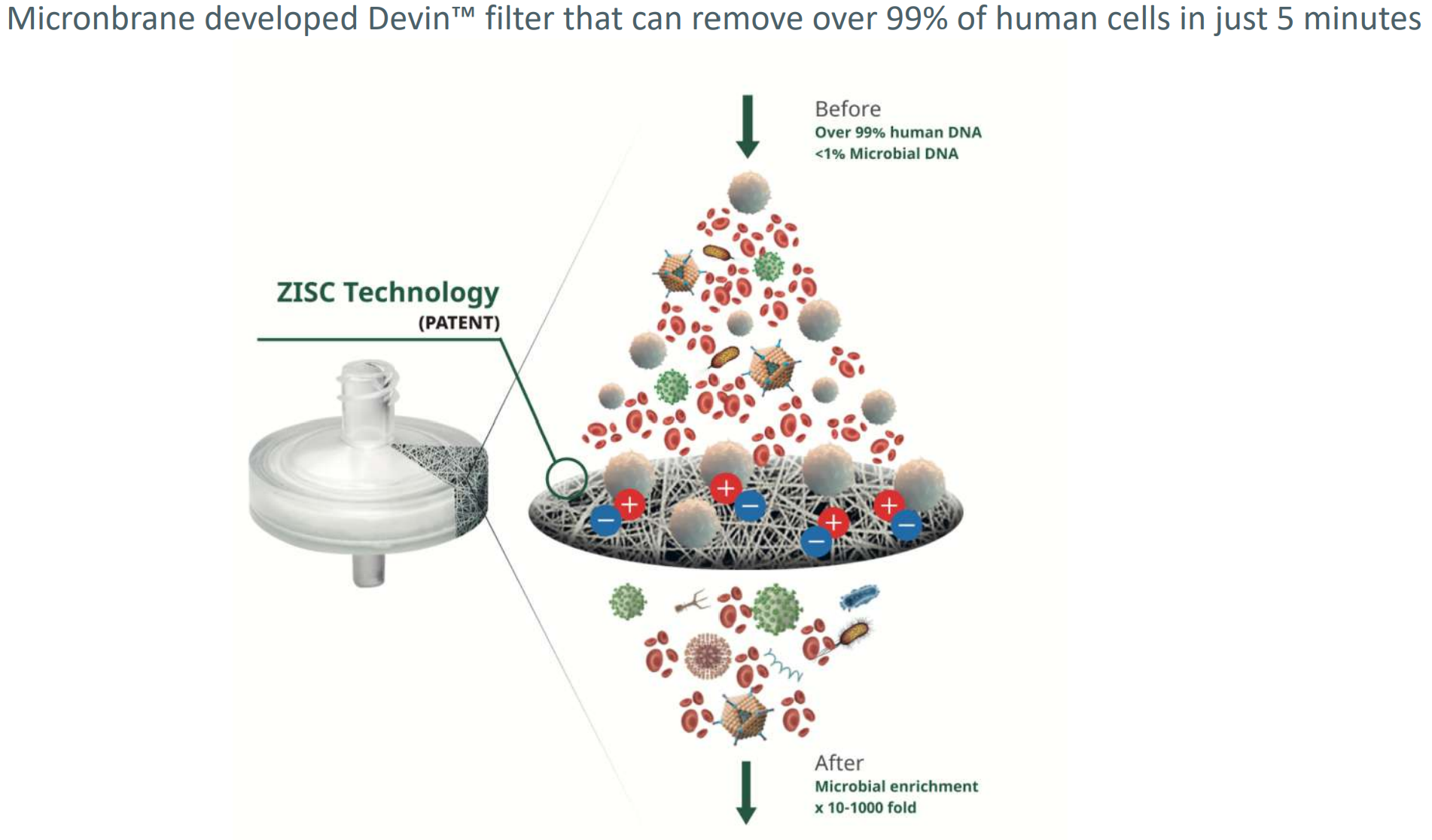
The barcterial DNA, Human DNA and Methylated DNA after elution will pass the filter while the lymphocyte debri will adhere to the Devin filter.
This is not a positive selection like with anion exchange DNA plasmid preps or mRNA extraction with charges silica glass that is used for microRNA profiling.
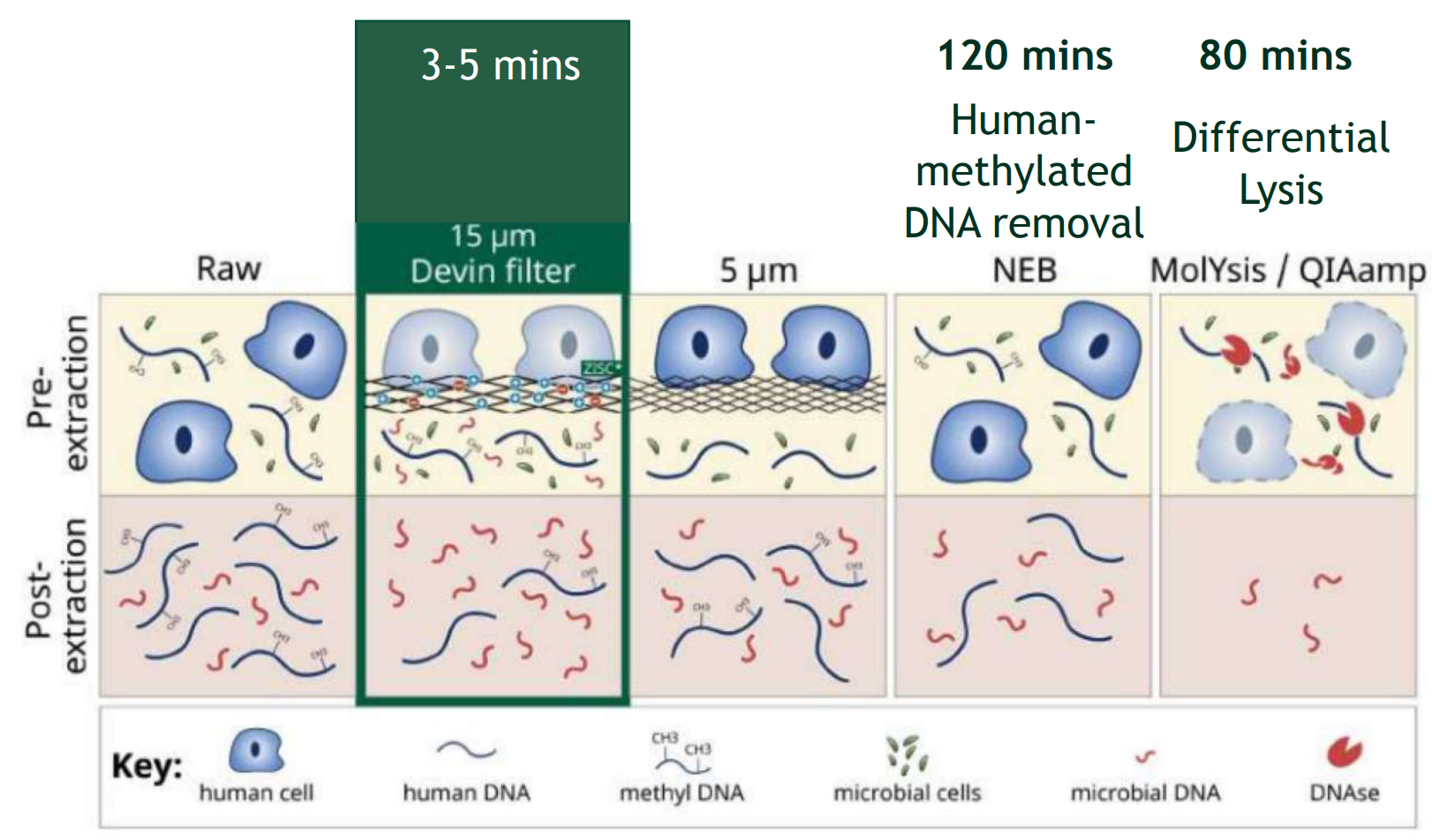
Zwitterionic Coating (ZISC) doesn’t rely on membrane pore size and effectively binds only to nucleated cells. In comparison with other methods (e.g. human methylated DNA removal, differential lysis) Devin™ filter doesn’t impact microbial DNA and leaves microorganisms completely intact .
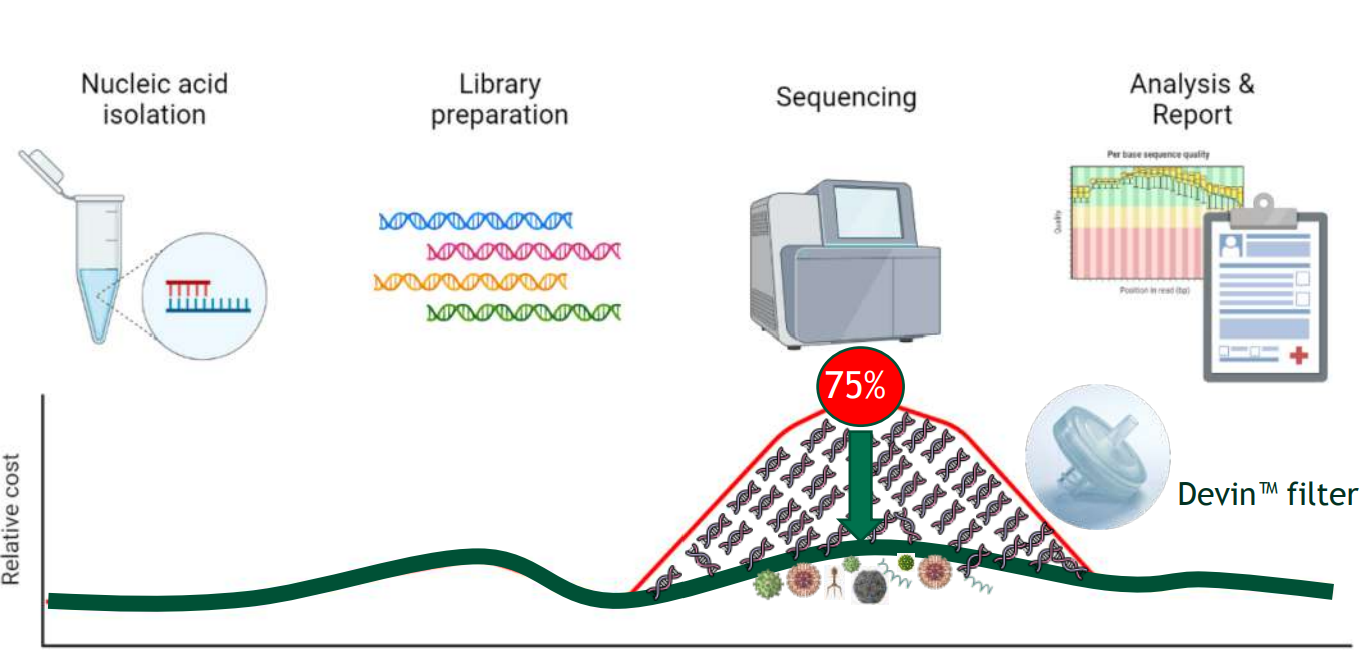
Devin™ filter is a one step sample preparation, doesn’t require special skills and is not time/labor consuming, it will take only 2 minutes!
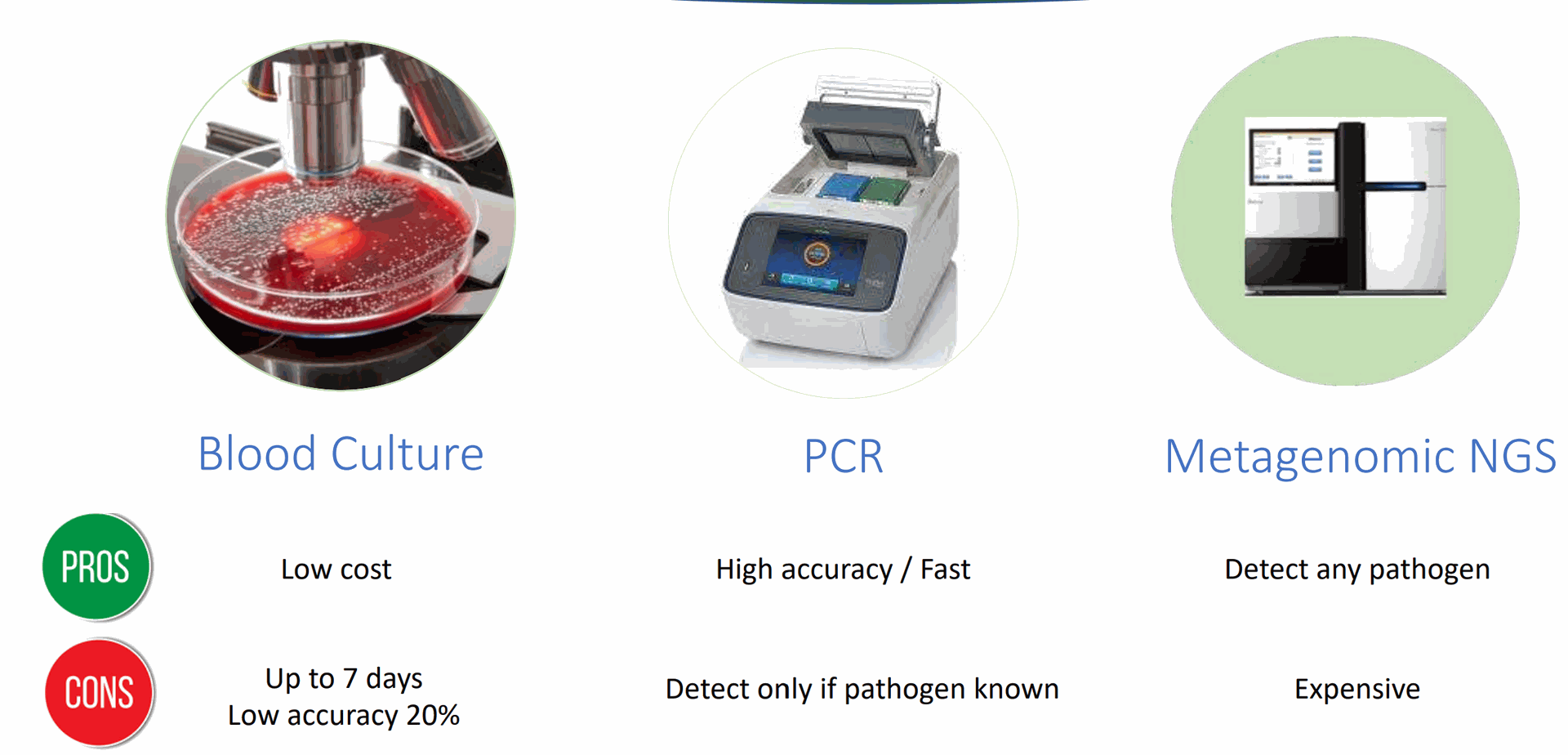
Lieven Gevaert, Bio-engineer at Gentaur has tested 3 systems to isolate bacterial DNA. Lieven Gevaert's Bio, Sales Gear and Research Gate

- Devin 5 minutes protocol is the best
- MolYsis QiaAmp 80 minutes came out as second
- NEB the New England Biolabs Human Methylated DNA removal kit 120 minutes came out at the 3th place.
Devin™ is a fractionation filter that effectively removes host DNA interference from body fluids, such as whole blood, plasma, CSF, BALF etc. It utilizes a patent technology of Zwitterionic Interface UltraSelf-assemble Coating (ZISC) which allows to effectively deplete human nucleated cells from the sample, while allowing most of microorganisms (including bacteria, viruses, fungi etc.) to go through the filter intact.
Due to high reduction of host DNA Devin™ enables to reduce 75% of downstream sequencing cost.
Buy Zitterion coated purification kits
|
Description |
Catalogue # |
1 box includes |
Unit Price, USD |
Transportation and Storage Conditions |
|
|
1 |
Devin™ Fractionation Syringe Filter |
DF-01-024 |
24 filters |
580.0 |
Transportation and storage under room temperature |
|
2 |
Devin™ Microbial DNA Enrichment Kit |
MEK-01-008 |
8 rxns |
340.0 |
Transportation at room temperature. When shipped, Store Proteinase K at 2°C~8°C and The Lysozyme at −25°C~−15°C (they come into separate boxes). |
|
3 |
Devin™ Microbial DNA Enrichment Kit |
MEK-01-024 |
24 rxns |
750.0 |
Transportation at room temperature. When shipped, Store Proteinase K at 2°C~8°C and The Lysozyme at −25°C~−15°C (they come into separate boxes). |
|
4 |
PaRTI-Seq™ Ultralow DNA NGS library preparation Kit |
NLK-01-008 |
8 rxns |
340.0 |
Transportation and storage at -20°C |
|
5 |
PaRTI-Seq™ Ultralow DNA NGS library preparation Kit |
NLK-01-048 |
48 rxns |
1500.0 |
Transportation and storage at -20°C |

Zwitterion isolation compared to Qiagen's MolYsis™ technology for bacterial and fungal DNA isolation from a human blood.
Using MolYsis™-extracted DNA, detection limits of PCR assays, such as those directed to 16S and 18S rRNA genes of bacteria and fungi, respectively, lie at 10-40 cfu/ml from Candida albicans or S. aureus or E. coli will show lower pathogen loads prevailing in the blood than Devin filtration with zwitterion coaded Polypropylene.
Processing of 5-10ml blood decreases the detection limit even more. So far, Devin extraction and real-time PCR with sequencing analysis identified species from 86 Gram-positive and 120 Gram-negative bacterial and 65 fungal genera.
Compared to the NEBNext® Microbiome DNA Enrichment Kit for enrichment of microbial DNA from samples containing methylated host DNA (including human)
Selective binding and removal of the CpG-methylated host DNA. Importantly, microbial diversity remains intact after enrichment.
Results
- Effective enrichment of microbial genomic DNA from samples contaminating host DNA
- Fast, simple but long protocol 120 min.
- Enables microbiome whole genome sequencing, even for samples with high levels of host DNA
- Compatible with downstream applications including next generation sequencing on all platforms, qPCR and end point PCR
- Suitable for a wide range of sample types
- No requirement for live cells
- Optional protocol to retain separated host DNA
- Also effective for separation of organelle DNA (e.g. mitochondria, chloroplast) from eukaryote nuclear DNA

1 DEVIN™M Fractionation
Syringe Filter DF-01-024 580 USD
2 DEVIN™ Microbial DNA
Enrichment Kit MEK-01-008 340USD
3 DEVIN™ Microbial DNA
Enrichment Kit MEK-01-024 750 USD
4 Unison™ Ultralow DNA NGS
library preparation Kit NLK-01-008 340 USD
5 Unison™ Ultralow DNA NGS
library preparation Kit NLK-01-048 1500USD
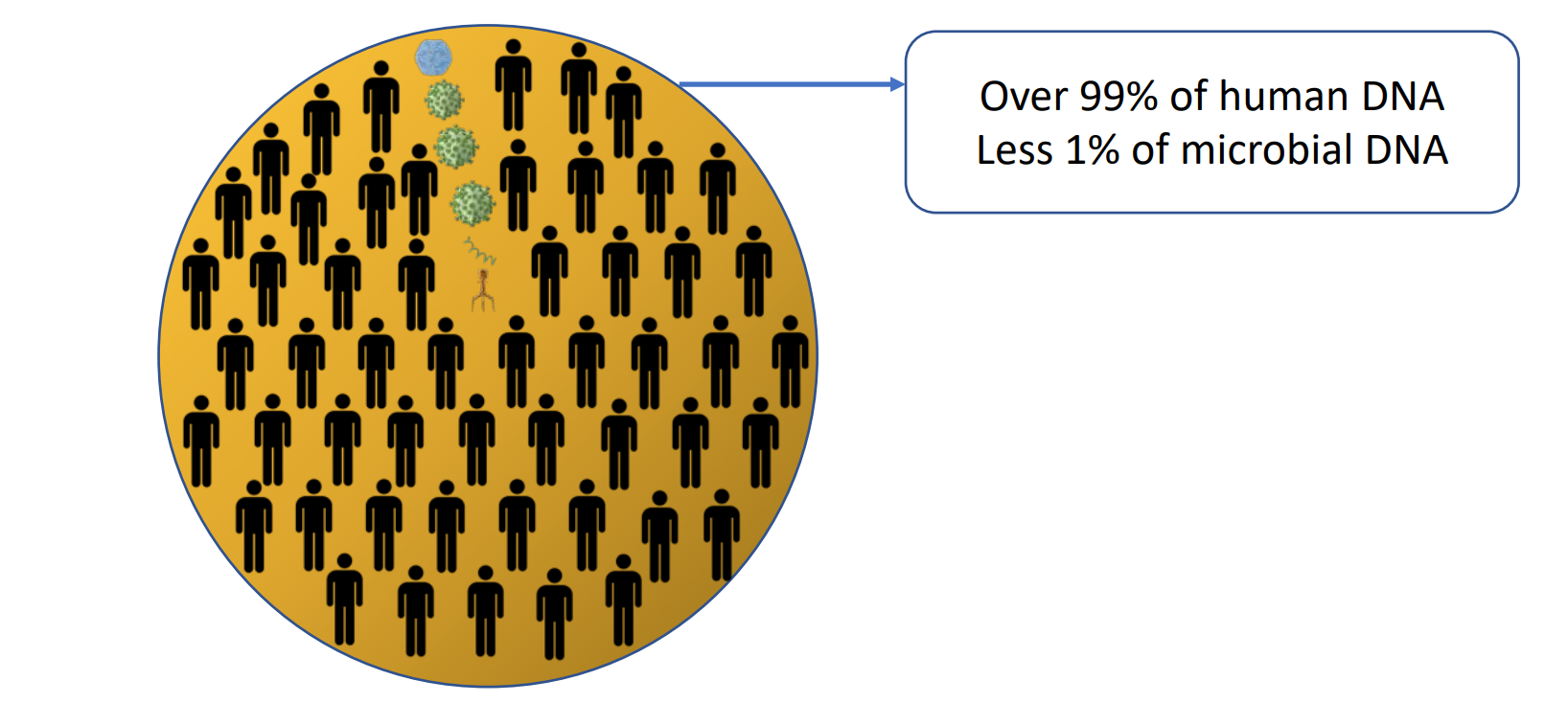
In human DNA, 4-6% of cytosines are methylated, and 60-90% of these methylated cytosines are at CpG sites .
Methylation at CpG sites in microbial species is rare.
The Zwitterion Microbiome DNA Enrichment Kit uses a simple and fast polypropylene filter method to selectively bind and remove lymphocytes. The method uses zwitterions to pull out the CpG-methylated (eukaryotic) DNA, leaving the non-CpG-methylated (microbial) DNA in the eluent.
The ondex Microbiome DNA Enrichment Kit is suitable for a wide range of sample types, including samples with high levels of contaminating host DNA, and is also effective for separation of organelle DNA (e.g. mitochondria, chloroplast) from eukaryote nuclear DNA 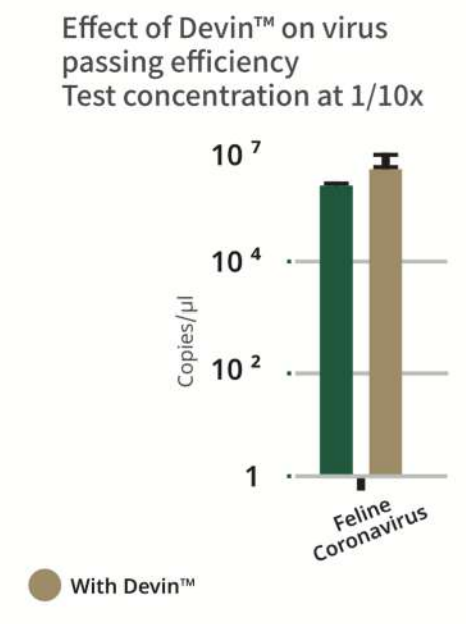
The Devin kit is compatible with downstream applications including next generation sequencing on all platforms, qPCR and end-point PCR.
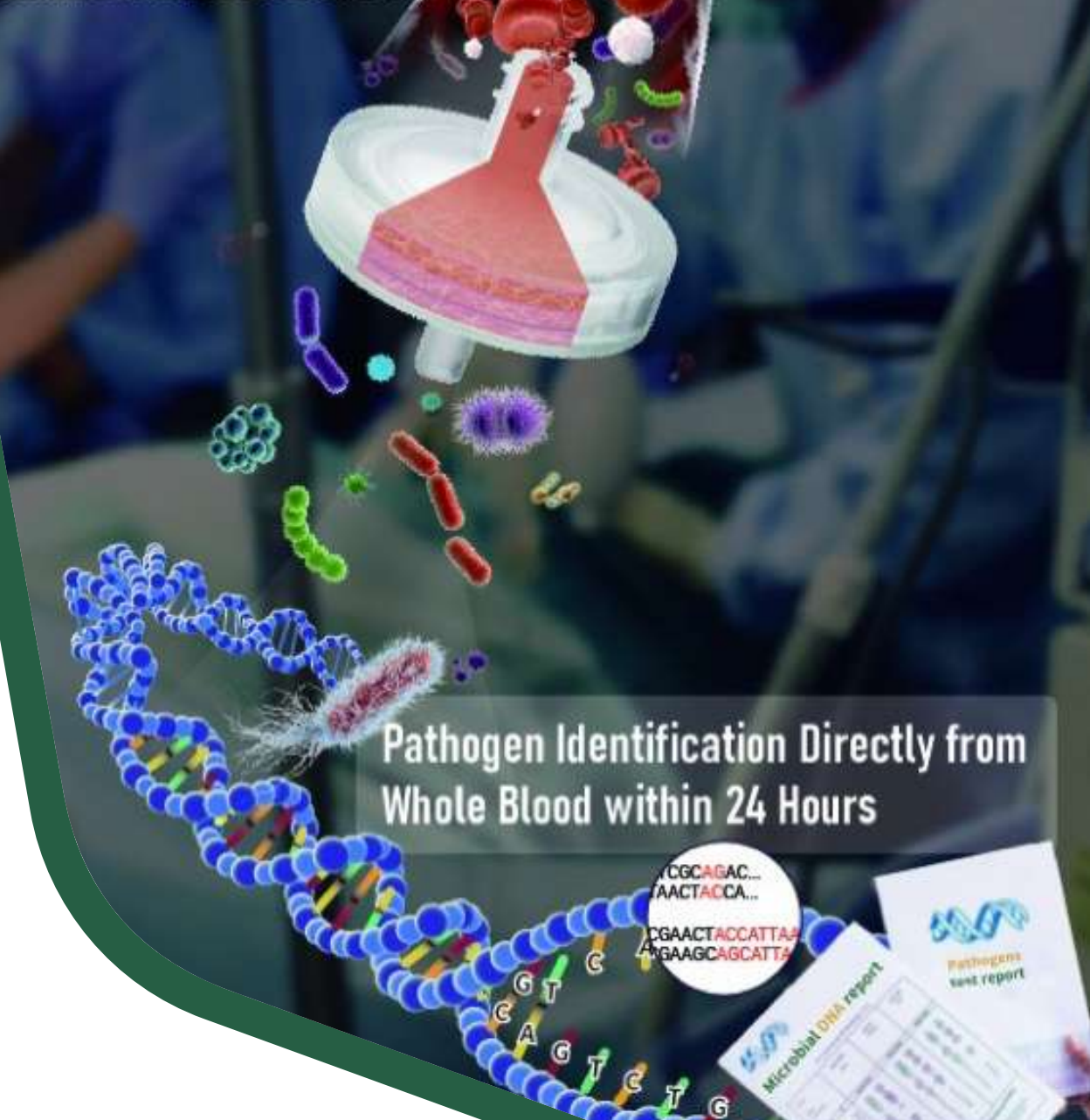
Based on Devin™ Micronbrane built the complete solution for pathogen identification which is called PaRTI-Seq™. PaRTI-Seq™ includes filter, consumables and reagents for optimized sample preparation, microbial DNA extraction, Sequencing and proprietary software for analysis to get precise test results within 24 hours.
Available for purchase!
n Includes Devin™ fractionation filters for host cells depletion
n Optimized extraction condition for both Gram- and Gram+ bacteria
n Fast and simple protocol (both manual and automated available)
n Compatible with downstream applications including next generation
sequencing on all platforms, qPCR and end point PCR
High Efficiency Microbial DNA
Enrichment Kit (24 reactions)
Optimized for fast and effective
extraction of microbial DNA from
sample
*Results of human and microbial identification using qPCR, NGS and Nanopore. Human
blood samples (5mL) were added 10*4 Genome Copies/mL spike-in control from ZYMO
Research and then processed using different depletion methods. Test results show that
Devin® filter increases ratio of microbial DNA and decreases the ratio of human (host) DNA.
Coupled with Devin™ filter microbial
enrichment can increase 10– 1000 fold with
much better results of microbial sequencing
reads vs other methods. Therefore it’s
possible to detect even tiny amount of
pathogens (102 genome copies/mL)

References
[1] Kellogg JA et al. (2000) J Clin Microbiol 38, 2181-
2185.
[2] Disqué C (2007) BIOspektrum 06, 627-629.
[3] Krohn S et al. (2014) J Clin Microbiol 52, 1754-1757.
[4] Peterson SW et al. (2016) PLoS ONE 11, e0152493.
doi:10.1371/journal.pone.0152493.
[5] Duran-Pinedo AE et al. (2014) The ISME Journal,
doi:10.1038/ismej.2014.23.
[6] Horz HP et al. (2008) J Microbiol Meth 72, 98-102.
[7] Leo S et al. (2017) Int J Mol Sci 18(9):2011,
doi:10.3390/ijms18092011.
[8] Thoendel M et al. (2016) J Microbiol Meth, doi:
10.1016/j.mimet.2016.05.022.
[9] Esteban J et al. (2012) Acta Orthopaedica 3, 299-304.
[10] Xu Y et al. (2012) FEMS Immunol Med Microbiol 65,
291–304.
[11] Schmidt K et al. (2017) J Antimicrob Chemother 72:
104–114, doi:10.1093/jac/dkw397
[12] Ruppé et al. (2017) Scientific Reports volume 7, Article
number: 7718, doi:10.1038/s41598-017-07546-5
[13] Rudkjøbing VB et al. (2014) J Cys Fibrosis
doi:10.1016/j.jcf.2014.02.008.
[14] Hansen WLJ et al. (2009) J Clin Microbiol 47, 2629-
2631.
[15] Zhao Y et al. (2014) J Clin Microbiol 52, 2212-2215.
[16] Benítez-Páez A et al. (2013) PLoS ONE 8, e57782.
doi:10.1371/journal.pone.0057782.
[17] Gyarmati P et al. (2015) PLoS ONE 10, e0135756.
doi:10.1371/journal.pone.0135756.
[18] Gyarmati P et al. (2016) Sci Rep. 6, 23532. doi:
10.1038/srep23532.
[19] Marín MJ et al. (2016) Med Oral Patol Oral Cir Bucal
21, e276-84. doi:10.4317/medoral.20842.
[20] Knabl L et al. (2015) J Microbiol Meth 120, 91-93.
[21] Dada N et al. (2018) Scientific Reports 8: 2084
[22] Arrazuria R et al. (2016) Front Microbiol 7, 446. doi:
10.3389/fmicb.2016.00446.
[23] Kleinschmidt K et al. (2017) J Appl Microbiol 122, 997-
1008.
[24] McCann CD, Jordan JJ (2014) J Microbiol Meth 99, 1-